Automating ETL Workflows with Apache Airflow: An Universal Solution for Data Management
Introduction
Vast about of data is being processed daily in various manners like financial transactions, customer interactions, sensors, research results, and so on. For instance, pharmaceutical companies produce millions of data points just from clinical trials alone. This huge amount of data has to be handled with accuracy and speed. Otherwise it can result in complications such as slow regulatory approvals and loss of business opportunities.
Revolutionizing Pharmaceutical Data Workflows with Apache Airflow
Pharmaceutical companies rely on massive amounts of information that must be carefully managed at several stages from the early research and development, clinical testing, and final regulatory submission. Good management and automation of such data processes ensures accuracy and standards, at the same time avoids costly delays in bringing new drugs to market.
Apache Airflow is an industry standard open-source ETL tool that helps in managing complicated workflow which makes it suitable for usage within the pharmaceutical industry as well. We will be showcasing an example of Airflow based development to help automate clinical trial data processing.
Business Use Case: Automated Clinical Trial Data Processing
A pharmaceutical company is running several clinical trials at the same time. The extent of data generated in each clinical trial is extensive. It requires systematic processing, cleaning, and analysis. The company intend to use an automated solution to drive ETL(Extract-Transform-Load) for clinical trial data management, thus ensuring timely and precise reporting to regulatory agencies and other stakeholders.
Problems:
- Data Volume and Complexity: Reconciliation of millions of data points without human intervention
- Compliance: Ensure accuracy of data and time-bound filing with different regulatory authorities
- Resource Allocation: Engage manual efforts spent on data processing to strategic activities
By automating the ETL process with Apache Airflow, the company can address these challenges effectively, improving operational efficiency, enhancing data accuracy, and ensuring compliance.
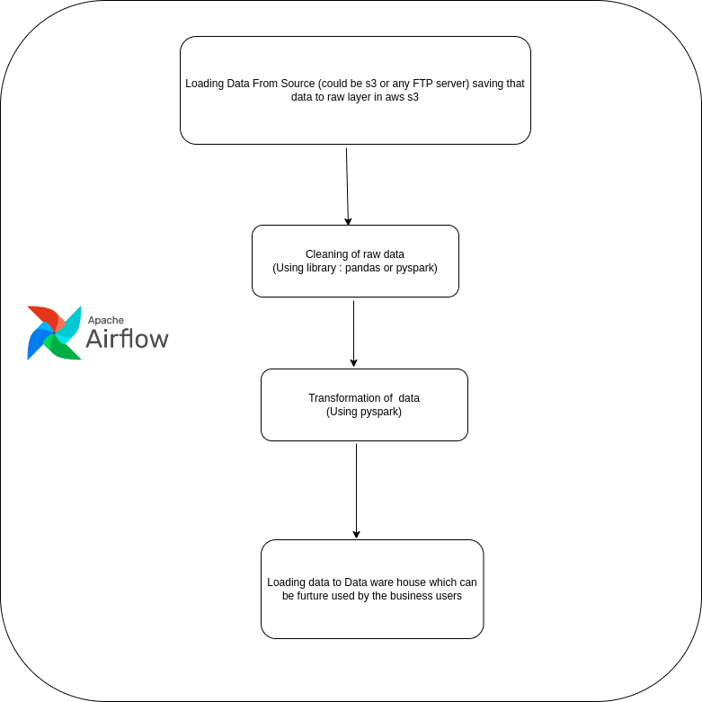
airflow-workflow-diagram
The above Diagram shows the workflow of processing the clinical data using airflow.
Now, let’s walk through how Apache Airflow can be used in automating the clinical trial data processing workflow. We’re going to create a dummy workflow (DAG) to illustrate steps to achieve this with example data.
Implementation of Clinical Trial Data Processing with Apache Airflow
What is Apache Airflow?
Apache Airflow is an open-source platform designed for programmatic authorization, scheduling, and monitoring of workflows. Users can create DAGs where each node is a task interconnected by edges, which define dependencies among nodes. Originally developed at Airbnb and handed over to the Apache Software Foundation, Airflow is currently used to help companies in various sectors to manage complex workflows.
Key Features of Apache Airflow
- Dynamic: Airflow workflows are defined in Python, hence allowing the generation of pipelines dynamically. This allows one to use complex logic for pipeline creation.
- Scalable: It can scale to handle an increasing number of tasks by using a distributed execution engine.
- Extensible: Airflow has an architecture designed for easy addition of new operators and sensors, hooks—making it highly customizable.
- UI: Airflow has a rich user interface so that pipelines can be visualized, progress can be tracked in real time, and issues can be debugged. Additionally, Airflow allows both the manual triggering of tasks and easy reruns of failed tasks.
- Scheduler: Airflow’s scheduler will run tasks in the appropriate order. Retries will be handled; workflows should run on time or when certain external events occur.
- Integrations: Airflow integrates with a wide variety of services and systems, including databases, cloud storage, data warehouses, and many others.
Prerequisite:
– Python should be installed on the system (python 3.7 or later version)
Step 1: Setting Up Airflow
First, install Apache Airflow using the following commands:
– pip install apache-airflow
– airflow db init
– airflow users create \
--username admin \
--firstname Admin \
--lastname User \
--role Admin \
--email admin@example.com
Additionally, install the necessary packages to interact with S3, Postgres, or Redshift
– pip install boto3 pandas psycopg2-binary
Start the Apache Airflow service
– airflow webserver --port 8080 – airflow scheduler
Step 2: Creating the Workflow (Dag)
A DAG is a Directed Acyclic Graph in Airflow, visualizing a workflow; every node represents a task, while edges define dependencies between tasks.
The following is a dummy DAG for the processing workflow of clinical trial data:
from airflow import DAG
from airflow.operators.dummy import DummyOperator
from airflow.operators.python import PythonOperator
from airflow.models import Variable
from datetime import datetime
import boto3
import pandas as pd
import psycopg2
#session
session = boto3.Session(
aws_access_key_id=Variable.get('Access key'),
aws_secret_access_key=Variable.get('Secret access key'),
)
# Configuration for S3 and PostgreSQL
S3_BUCKET_NAME = 'airflow-demo-bucket1'
S3_KEY = 'source/clinical_data.csv'
CLEANED_DATA_KEY = 'source/cleaned_data/cleaned_clinical_data.csv'
ANALYSIS_SUMMARY_KEY = 'path/to/clinical_analysis_summary.csv'
POSTGRES_HOST = 'localhost'
POSTGRES_PORT = '5432'
POSTGRES_DB = 'clinicTrial'
POSTGRES_USER = 'prince'
POSTGRES_PASSWORD = '12345'
POSTGRES_TABLE = 'clinical_trial_data'
def clean_column(df, column_name):
for x in df.index:
if df.loc[x, column_name] == '.' or df.loc[x, column_name] == ' ':
df.loc[x, column_name] = None
def extract_clinical_data():
# Extract clinical trial data from S3
print("Extracting clinical trial data from S3...")
s3 = session.client('s3')
response = s3.get_object(Bucket=S3_BUCKET_NAME, Key=S3_KEY)
clinical_data = pd.read_csv(response['Body'])
clinical_data.to_csv('/tmp/clinical_data.csv', index=False)
print("Data extraction complete.")
def clean_clinical_data():
# Clean clinical trial data
print("Cleaning clinical trial data...")
clinical_data = pd.read_csv('/tmp/clinical_data.csv')
clinical_data['Specimen_date'] = clinical_data['Specimen_date'].str.strip()
clinical_data['Specimen_date'] = pd.to_datetime(clinical_data['Specimen_date'], format='mixed')
clean_column(clinical_data, "Date_of_Death")
clinical_data['Date_of_Death'] = pd.to_datetime(clinical_data['Date_of_Death'], format='mixed')
clinical_data['Date_of_Last_Follow_Up'] = pd.to_datetime(clinical_data['Date_of_Last_Follow_Up'], format='mixed')
clinical_data['Time'] = pd.to_numeric(clinical_data['Time'])
clinical_data['Event'] = clinical_data['Event'].astype(int)
clinical_data.to_csv('/tmp/cleaned_clinical_data.csv', index=False)
# Upload the cleaned data to S3
s3 = session.client('s3')
s3.upload_file('/tmp/cleaned_clinical_data.csv', S3_BUCKET_NAME, CLEANED_DATA_KEY)
print("Data cleaning complete.")
def analyze_clinical_data():
# Analyze clinical trial data
print("Analyzing clinical trial data...")
s3 = session.client('s3')
s3.download_file(S3_BUCKET_NAME, CLEANED_DATA_KEY, '/tmp/cleaned_clinical_data.csv')
cleaned_data = pd.read_csv('/tmp/cleaned_clinical_data.csv')
# Calculate survival time for each patient
cleaned_data['Specimen_date'] = pd.to_datetime(cleaned_data['Specimen_date'], format='mixed')
cleaned_data['Date_of_Death'] = pd.to_datetime(cleaned_data['Date_of_Death'], format='mixed')
cleaned_data['Survival_Time'] = (cleaned_data['Date_of_Death'] - cleaned_data['Specimen_date']).dt.days
summary = cleaned_data.groupby('Stage')['Survival_Time'].mean()
summary.to_csv('/tmp/clinical_analysis_summary.csv')
# Upload the analysis summary to S3
s3.upload_file('/tmp/clinical_analysis_summary.csv', S3_BUCKET_NAME, ANALYSIS_SUMMARY_KEY)
print(f"Data analysis complete. Summary:\n{summary}")
def load_data_to_postgres():
# Load data into PostgreSQL
print("Loading data into PostgreSQL...")
s3 = session.client('s3')
s3.download_file(S3_BUCKET_NAME, CLEANED_DATA_KEY, '/tmp/cleaned_clinical_data.csv')
cleaned_data = pd.read_csv('/tmp/cleaned_clinical_data.csv')
cleaned_data.dropna(inplace=True)
conn = psycopg2.connect(
dbname=POSTGRES_DB,
user=POSTGRES_USER,
password=POSTGRES_PASSWORD,
host=POSTGRES_HOST,
port=POSTGRES_PORT
)
cursor = conn.cursor()
# Create table if not exists (for simplicity)
create_table_query = f"""
CREATE TABLE IF NOT EXISTS {POSTGRES_TABLE} (
PatientID INT,
Specimen_date DATE,
Dead_or_Alive VARCHAR(5),
Date_of_Death DATE,
Date_of_Last_Follow_Up DATE,
sex VARCHAR(1),
race VARCHAR(1),
Stage VARCHAR(10),
Event INT,
Time INT
);
"""
cursor.execute(create_table_query)
# Insert data into the table
for _, row in cleaned_data.iterrows():
insert_query = f"""
INSERT INTO {POSTGRES_TABLE} (PatientID, Specimen_date, Dead_or_Alive, Date_of_Death, Date_of_Last_Follow_Up, sex, race, Stage, Event, Time)
VALUES ({row['PatientID']}, '{row['Specimen_date']}', '{row['Dead_or_Alive']}', '{row['Date_of_Death']}', '{row['Date_of_Last_Follow_Up']}', '{row['sex']}', '{row['race']}', '{row['Stage']}', {row['Event']}, {row['Time']});
"""
cursor.execute(insert_query)
conn.commit()
cursor.close()
conn.close()
print("Data loaded into PostgreSQL.")
default_args = {
'owner': 'airflow',
'depends_on_past': False,
'start_date': datetime(2024, 7, 9),
'retries': 1,
}
with DAG('clinical_trial_data_processing', default_args=default_args, schedule_interval='@daily', catchup=False) as dag:
start = DummyOperator(task_id='start')
extract = PythonOperator(
task_id='extract_clinical_data',
python_callable=extract_clinical_data
)
clean = PythonOperator(
task_id='clean_clinical_data',
python_callable=clean_clinical_data
)
analyze = PythonOperator(
task_id='analyze_clinical_data',
python_callable=analyze_clinical_data
)
load = PythonOperator(
task_id='load_data_to_postgres',
python_callable=load_data_to_postgres
)
end = DummyOperator(task_id='end')
start >> extract >> clean >> analyze >> load >> end
Step 3: Running the Workflow
- Create a dags folder and save the DAG file as clinical_trial_data_processing_dag.py in the dags directory of your Airflow installation.
- Start the Airflow webserver and scheduler if they are not already running
- airflow webserver –port 8080
- airflow scheduler
- Access the Airflow web UI by navigating to http://localhost:8080 in your web browser.
- Enable the clinical_trial_data_processing DAG from the web UI and monitor its execution.
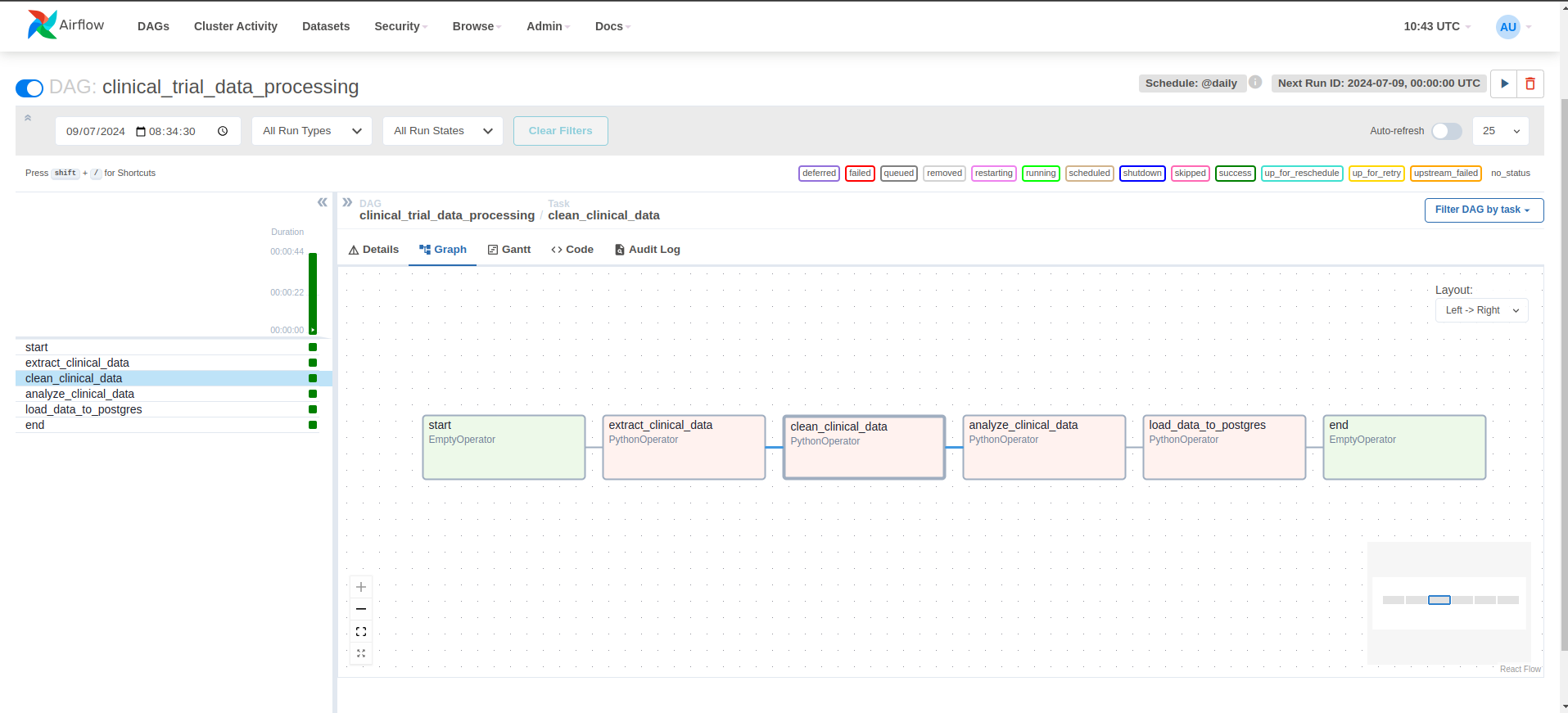
airflow-workflow-web-ui.png
Benefits gained by the Business:
Apache Airflow aids pharmaceutical companies to be able to:
- Enhance Efficiency: Automate repetitive tasks and free up some of the most valuable resources
- Improved Accuracy: Reduce human errors in data processing
- Compliance: Ensure that reports are submitted on time and with accuracy to various regulatory bodies
- Scalability: Handle increasing volumes of data with ease as clinical trials grow in size
Conclusion
Apache Airflow is positioned to help pharmaceutical companies transform data management from an overwhelming task into a streamlined system. It should be an important component in a company’s Digital Transformation Toolkit to drive innovation and achieve operational efficiency.

